Welcome to the FESNov catalog explorer
A substantial portion of Earth's microbes remains uncultured and under-studied. This limited understanding extends to the functional and evolutionary aspects of their genetic material, which remains untraceable using current databases and overlooked in most metagenomic studies.
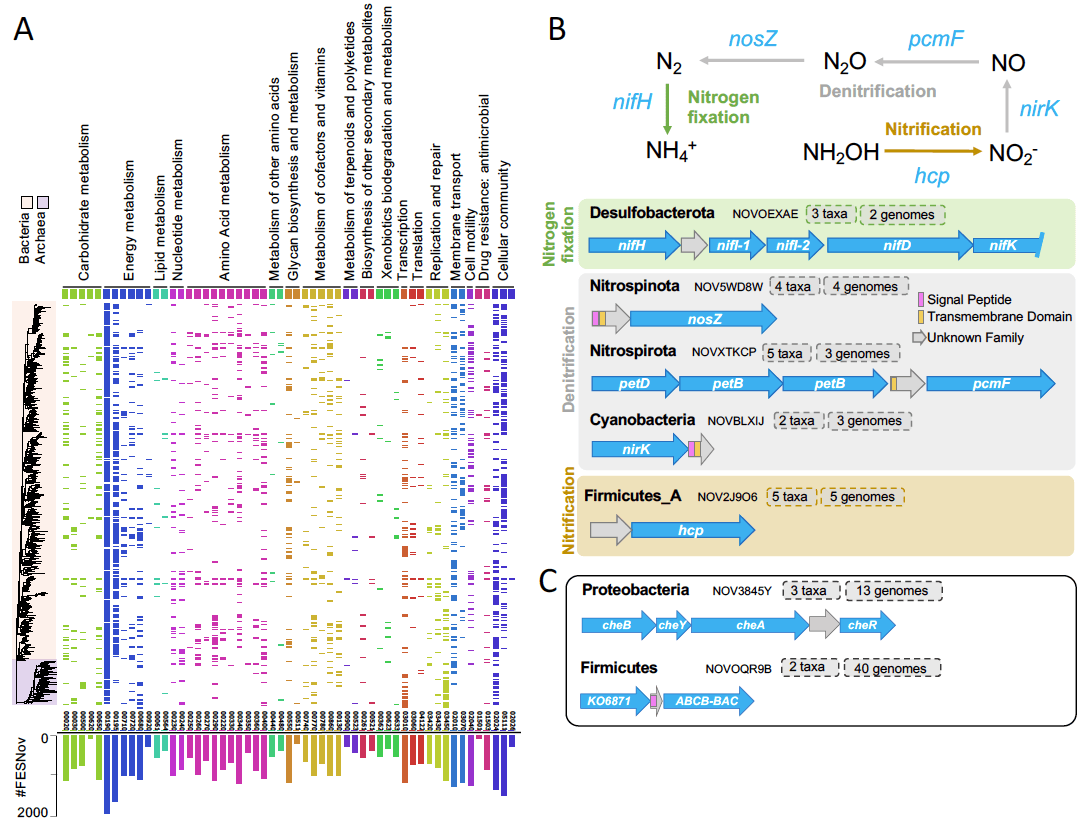
We analyzed 149,842 environmental genomes from multiple habitats and compiled a meticulously curated catalog of 404,085 functionally and evolutionarily significant novel (FESNov) gene families that are unique to uncultivated prokaryotic taxa.
All FESNov families span multiple species, exhibit strong signals of purifying selection and qualify as new orthologous groups, thus nearly tripling the number of bacterial and archeal gene families described to date.
The FESNov catalog is enriched in clade-specific traits, including 1,034 novel families that can precisely distinguish entire uncultivated phyla, classes, and orders, likely representing synapomorphic characteristics that facilitated their evolutionary divergence.
Using genomic context analysis and structural alignments, we established functional associations for 32.4% of the FESNov families, yielding 4,349 high-confidence associations with biological processes such as energy production, xenobiotic degradation, signaling, and antimicrobial resistance. These predictions provide a valuable hypothesis driven framework.
You can use this online browser to query FESNov gene families associated with specific KEGG pathway or including specific taxa.
| [[header.h_name]] |
|---|
| [[ entry[index] ]] |